pyAMNESIA
Published:
pyAMNESIA is a python pipeline for analysing the Activity and Morphology of NEurons using Skeletonization and other Image Analysis techniques.
Théo Dumont and I developed this python toolbox during a research internship at INMED, Cossart lab as part of the 2020 CENTURI internship program. For more information, you can check pyAMNESIA’s documentation and pyAMNESIA’s repository.
Motivation
In the context of studying the mechanisms by which hippocampal assemblies evolve during the development of mice (5 to 12 days), we dispose of some two-photon calcium imaging videos, such as this one:
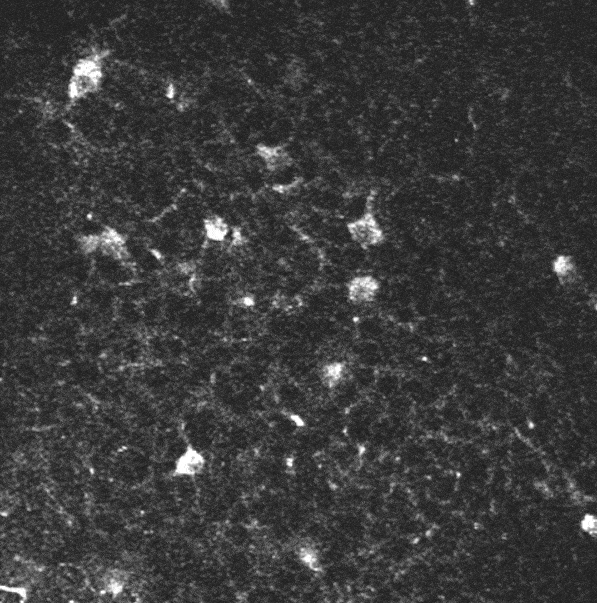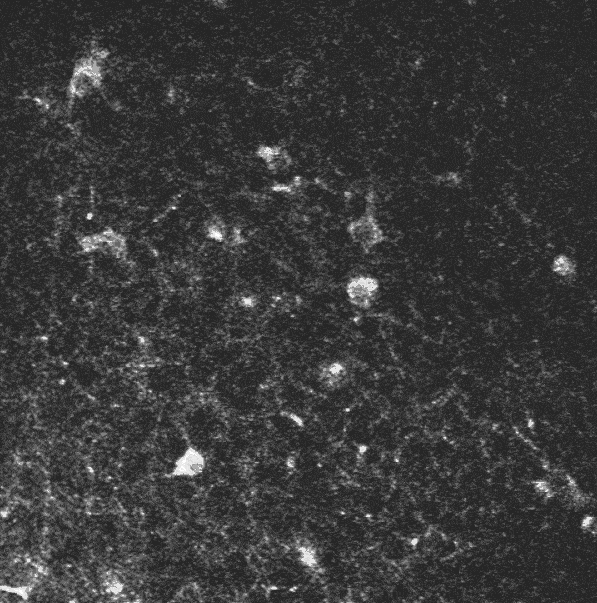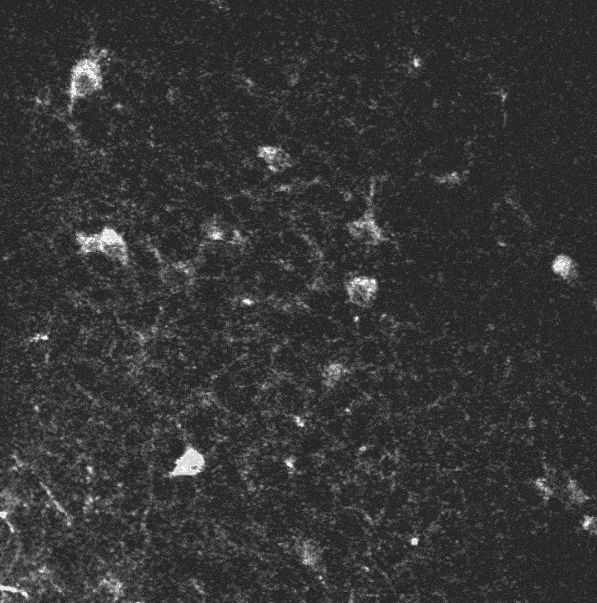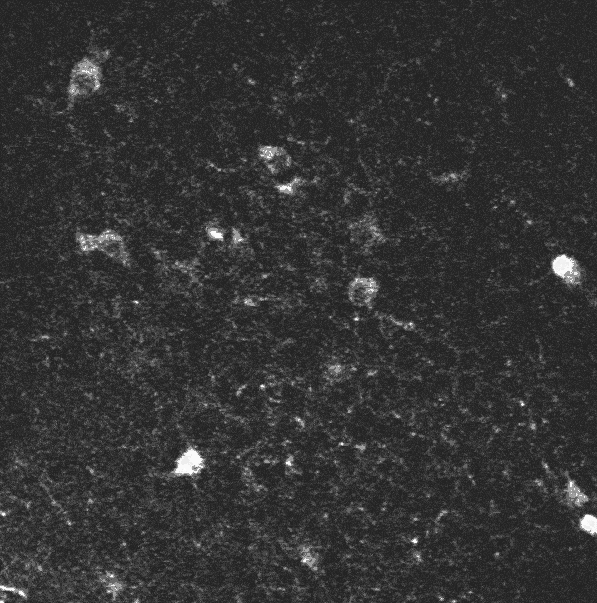
A two-photon calcium imaging movie.
The analysis of these calcium imaging videos may be split into two different parts:
- the evolution of the morphology, being the number of elements (somata, neurites…) and their visible connexions;
- the evolution of the neuronal activity, being the transients characteristics and the coactivity of the aforementioned elements.
In the state-of-the-art calcium imaging pipelines such as Suite2p and CaImAn , somata are segmented to subsequently study the evolution of their activity. In such cases, pixels in the region of a soma can reliably be considered as coactive, making the soma a morphological and functional entity.
Here, we want to study the whole neural structure of the hippocampus; not only the somata but also the neurites linking them. And there is no obvious correlation between the morphological “branches” that one can see on the videos, and the neurites, that are coactive functional entities. This is mainly due to the Z-axis projection - which “hides” vertical neurites and creates overlaps - but also to imaging hazards, or simply neurites that only activate partially during a neural transmission. Therefore, we do not know a priori whether a visible “branch” is a functional entity or not. We thus need to separate the morphological and the functional approaches, and try to build some coherent structures that could be considered as entities, in order to analyse their coactivity later.
Here is the problem we are trying to answer:
How can we use calcium imaging to get statistics on the evolution of interneurons in the hippocampus (morphology and activity) during mice development?
- input: a calcium imaging
.tifsequence - output: clusters of coactive pixels & morphological statistics
Functionalities
We split our approach in three parts:
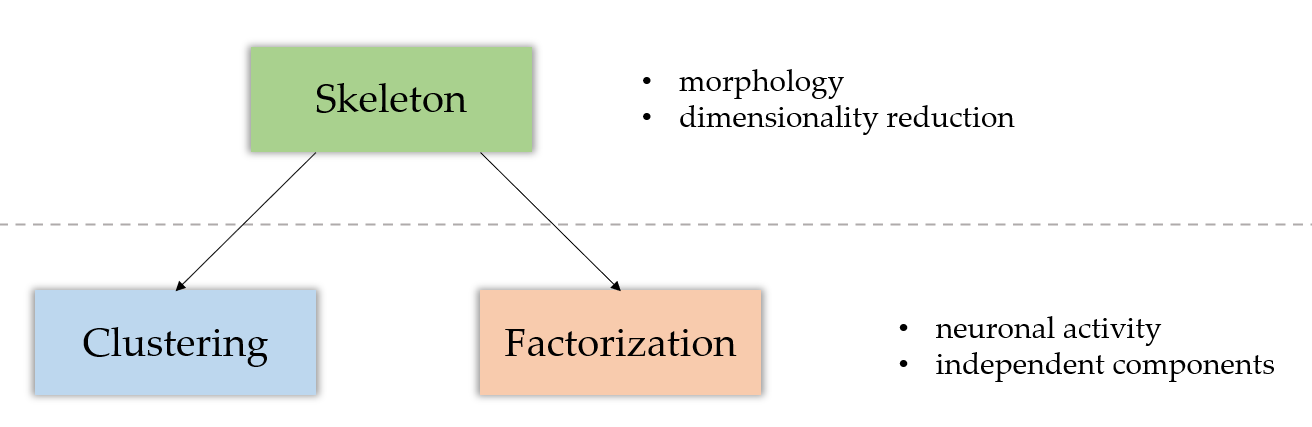 Structure of the approach
Structure of the approach
- The
skeletonizationmodule focuses on the morphological analysis of the data, by computing the underlying morphological skeleton of the sequence. - The
clusteringmodule performs an activity analysis of the elements in the sequence, whether they be pixels (no prior skeleton analysis) or branches (see the module page for more information about this). Its goal is to return clusters of coactive pixels. - The
factorizationmodule has a similar goal to the clustering module. It returns independent components of coactive pixels, but uses matrix factorization techniques to do so.
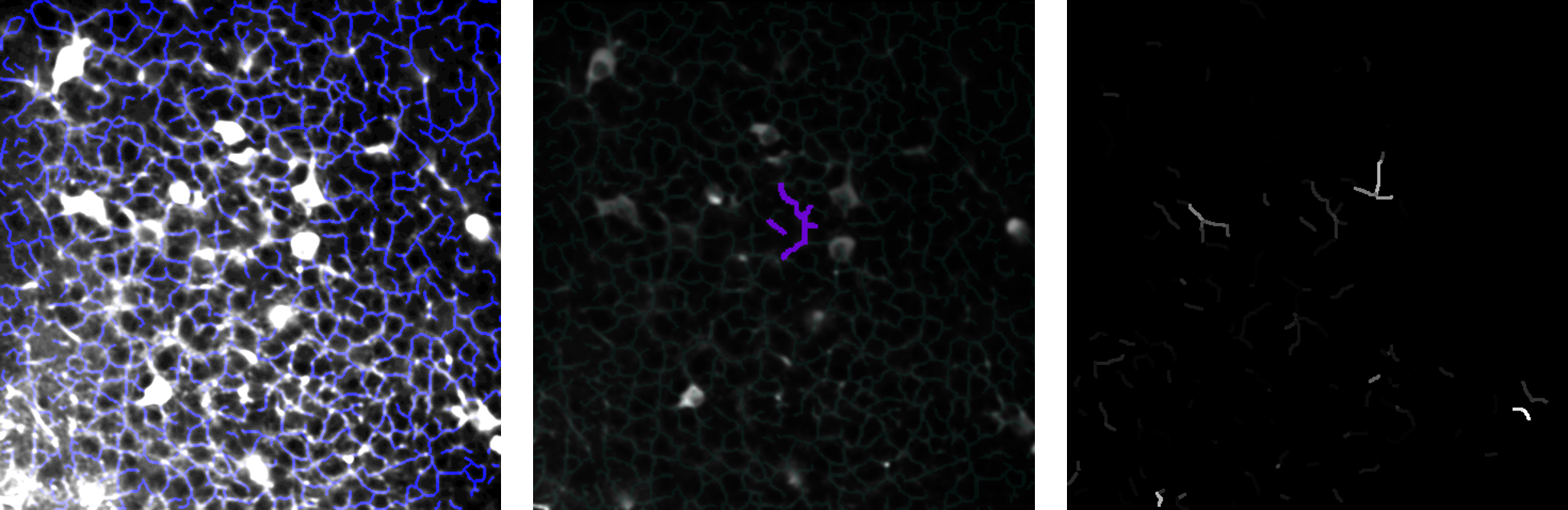 From left to right: a skeletonized image; a cluster; a factorization component
From left to right: a skeletonized image; a cluster; a factorization component
Whereas the skeletonization part focuses on the morphological aspect of the analysis, both of the clustering and factorization modules tackle the activity analysis of it. As shown on the diagram above, the skeleton module is prior to the two others.
For more information, you can check pyAMNESIA’s documentation and pyAMNESIA’s repository.
